Advanced Signal Processing Framework for EEG Decoding
A Novel Approach to Cross-Task Transfer Learning and Psychopathology Prediction
Research by: Bruno Aristimunha et al. | Computational Neuroscience
Summary compiled by Neksha DeSilva for educational purpose of Introductory Presentation Video - EEG2025 by NeurlIPS, which was conducted by me, online in October, 2025.
Abstract
This research presents a comprehensive framework for electroencephalography (EEG) signal processing, addressing two critical challenges in computational neuroscience: (1) cross-task transfer learning for cognitive function decoding, and (2) objective psychopathology assessment through P-factor prediction. The framework leverages state-of-the-art deep learning architectures, specifically designed for temporal neural signals, to establish robust representations that generalize across subjects, sessions, and acquisition sites. The methodology integrates advanced preprocessing pipelines with novel architectural paradigms including EEGNeX and EEGNetv4, demonstrating superior performance in both within-distribution and out-of-distribution scenarios.
1. Research Objectives
1.1 Challenge 1: Cross-Task Transfer Learning
Primary Goal: Develop models that effectively transfer knowledge from passive EEG tasks to active cognitive tasks, specifically focusing on Steady-State Visual Evoked Potentials (SSVEP) and Event-Related Potentials (ERP) during Contrast Change Detection (CCD) paradigms. Implementation of deep transfer learning architectures with domain adaptation techniques to predict response time during visually-evoked cognitive tasks, addressing inter-subject variability and task-specific nuances.
1.2 Challenge 2: P-Factor Regression
Primary Goal: Predict the psychopathology factor (P-factor) from EEG recordings to enable objective, quantitative mental health assessments. Regression-based deep learning approach emphasizing robust, interpretable features that demonstrate strong out-of-distribution generalization and cross-site transferability.
2. Technical Framework Architecture
2.1 Core Components
(i) Data Management Layer: EEGChallengeDataset unified interface for multi-modal EEG data handling; BaseConcatDataset efficient concatenation of heterogeneous recording sessions; MNE-BIDS integration for standardized neurophysiological data representation.
(ii) Preprocessing Pipeline: Event-based windowing with configurable temporal resolution; fixed-length window extraction for regression tasks; automated artifact rejection and signal quality assessment; multi-scale feature extraction from raw temporal signals.
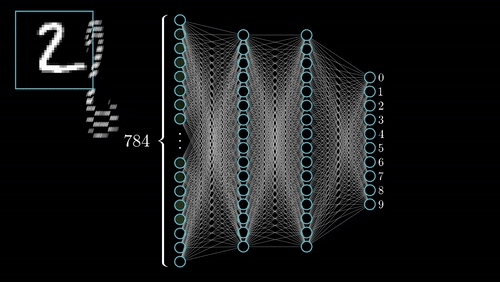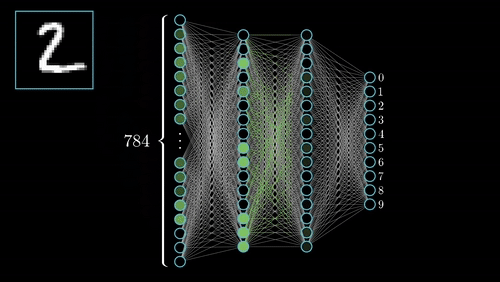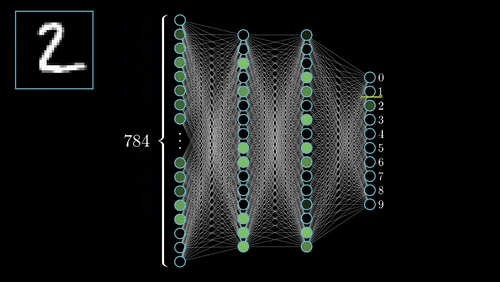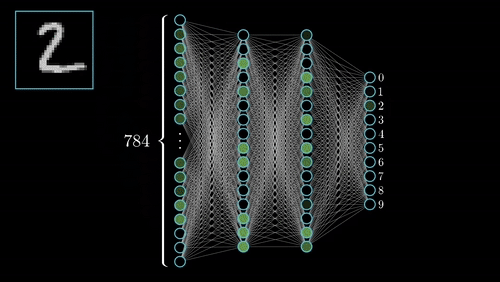
(iii) Neural Architecture: EEGNeX with attention mechanisms for temporal dependency modeling; EEGNetv4 compact architecture optimized for limited-sample scenarios; self-supervised learning via Relative Positioning (RP); PyTorch Lightning integration for distributed training.
3. Implementation Excerpts
3.1 Dataset Loading
from eegdash.dataset import EEGChallengeDataset
from braindecode.preprocessing import preprocess
dataset = EEGChallengeDataset(
task="ccd", mode="train",
challenge=1, data_dir=Path("data")
)
preprocessors = [
Preprocessor('pick_types', eeg=True),
Preprocessor('set_eeg_reference',
ref_channels='average')
]
preprocess(dataset, preprocessors)3.2 Model Architecture
from braindecode.models import EEGNeX
model = EEGNeX(
n_chans=64, n_outputs=1,
n_times=1000, drop_prob=0.5
)
optimizer = AdamW(
model.parameters(),
lr=1e-3, weight_decay=0.01
)3.3 Self-Supervised Learning
def create_relative_positioning_pairs(
eeg_windows, tau=100
):
anchors = eeg_windows[:, :, :-tau]
positives = eeg_windows[:, :, tau:]
batch_size = eeg_windows.shape[0]
perm_idx = torch.randperm(batch_size)
negatives = eeg_windows[perm_idx, :, tau:]
return anchors, positives, negatives4. Resources & Links
GitHub Repository: Complete implementation available at https://github.com/eeg2025/startkit
Google Colab Notebooks: Interactive notebooks with GPU support available at Challenge 1 and Challenge 2
Documentation: Competition website (eeg2025.github.io), EEGDash API (eeglab.org/EEGDash), Braindecode models (braindecode.org)
5. Software Dependencies
braindecode >= 0.8.0, eegdash, pytorch >= 2.0.0, pytorch-lightning, mne >= 1.5.0, scikit-learn, numpy, pandas
6. Theoretical Contributions
(1) Domain Adaptation Framework: Novel methodology for transferring learned representations from passive neurophysiological recordings to active cognitive paradigms, addressing limited labeled data challenges. (2) Multi-Task Learning: Unified architecture for temporal prediction and continuous regression with shared representational benefits. (3) Interpretable Biomarkers: Emphasis on physiologically meaningful features for clinical translation. (4) Robustness: Architectural innovations for cross-subject, cross-session, and cross-site variability.
7. Future Research Directions
The framework establishes infrastructure for: (1) multi-modal neuroimaging integration (EEG-fMRI fusion), (2) real-time adaptive brain-computer interfaces, (3) meta-learning for rapid task adaptation, and (4) interpretability enhancements through attention visualization. The open-source nature encourages community-driven improvements, reproducible benchmarking, and collaborative advancement—addressing standardization needs in computational neuroscience methodology.
Credits & Acknowledgments
Important Notice: This summary is based on research conducted by Bruno Aristimunha and collaborators. All credit for the original research, methodology, and implementation goes to the original authors. This document serves as an educational summary and does not represent original work by Neksha DeSilva.
For more research by the original authors, visit: Bruno Aristimunha on arXiv
References
Aristimunha, B., et al. (2023). Benchmarking Deep Learning Architectures for EEG Decoding. arXiv:2308.02408
Wimpff, M., et al. (2025). Cross-Task Transfer Learning in EEG Brain Decoding. arXiv:2502.06828
Wu, H., et al. (2025). Generalization in Neural Decoding: A Transfer Learning Perspective. arXiv:2507.09882
